PCR with Taq Polymerase
Procedure
- Prepare your template sample
- For amplifying Sinorhizobium sequences, boil a dense suspension of cells in 35 μL PCR lysis buffer for ~2 min and vortex
- For amplifying E. coli with Taq polymerase (but not Pfx50 polymerase), adding cells directly to the reaction is sufficient
- Thaw the Taq buffer, dNTPs, and primers
- Keep Taq polymerase on ice throughout the procedure
- Combine components according to one of the recipes below
- Set the reaction up on ice
- Select the appropriate cycling program and verify that all of the parameters are correct
- Allow 1 minute of extension time per 1 KB DNA being amplified (15-30 seconds is sufficient for Q5)
- Select "Run program" and select "YES" when asked about heated lid
- Wait until block has reached at least 70°C (this takes a little over 2 min)
- Place PCR tube into the machine
- Lower the lid until it latches and slightly tighten the heated lid onto the tubes
NOTE: When the PCR machine has finished cycling, your samples may be stored at -20°C. You may now proceed to gel electrophoresis and/or PCR clean-up.
Reaction recipes
20 μL
Taq
reactions
Use this for diagnostic purposes.

40 μL
Taq
reactions
This is the standard recipe. Use it in most cases for fragment amplification and sequencing.

40 μL reaction with
Pfx
polymerase
Use this for high-fidelity cloning.

20 μL reaction with
Q5
polymerase
Use this for high-fidelity cloning, especially for products >3kb.

40 μL reaction with
Q5
polymerase
Use this for high-fidelity cloning, especially for products >3kb.
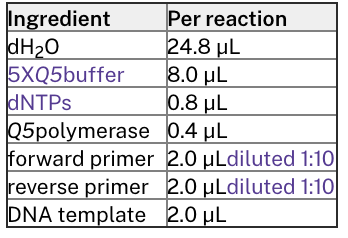
Master Mix Calculations

Primer Dilution
- 27 μL dH2O
- 3 μL primer
Repeat for both primers